From lab to production: streamlining biotech R&D
Abstract
Biotech is undergoing a transformation. Breakthroughs in omics, AI, and automation are accelerating discovery - but turning promising ideas into production-grade tools remains one of the biggest bottlenecks in science. At DeepLife, we’ve built a platform and culture that closes this gap.
Too often, research lives in a different universe from engineering. Scientific ideas get lost in Jupyter notebooks, deployments require handoffs and rewrites, and promising work stalls because the infrastructure isn’t ready. We believe there’s a better way.
In this blog, we share how we’ve operationalized R&D at DeepLife through a structured monorepo, modular components, and a fully automated stack - empowering scientists and engineers to work together in the same environment, with the same tools, at the same speed. We’ll take you inside our platform, philosophy, and real-world results.
We’re not just shipping code. We’re building the future of computational biology - where every discovery is one commit away from production.
The Challenge: Research and Production speak different languages
Scientific research thrives on curiosity and iteration. It is inherently exploratory, nonlinear, and messy - qualities that are essential to discovery, but notoriously difficult to integrate into scalable engineering systems. Meanwhile, production environments demand stability, security, observability, and predictability.
Too often, biotech companies are forced to choose between these two modes. Research code lives in ad-hoc notebooks or isolated GitHub repositories, while production code is treated as a separate world altogether. This creates silos, hinders reproducibility, and slows down the transfer of scientific insight into real-world applications.
At DeepLife, we’ve resolved this tension by building a single system that respects both needs. We’ve made it possible for researchers to remain flexible and autonomous, while still aligning with engineering principles that guarantee reliability at scale.
Potion: The core of DeepLife’s Engineering culture
The foundation of our approach is a structured monorepo we call Potion—short for Predictive Omics Tool for Indication and Off-target Narrowing. Potion captures the spirit of our work: part rigorous science, part alchemy. It reflects the dual nature of research: rigorous and reproducible on one hand, yet exploratory and adaptive on the other.
Potion isn’t just a code repository. It’s an ecosystem that enables collaboration across teams, supports modular code development, and enforces best practices by design. Built for both machine learning and bioinformatics, it serves as the unified backbone for our R&D workflows—from exploratory prototypes to production-grade pipelines.
The monorepo model offers several critical advantages. It enables atomic commits across projects, centralizes dependency management, and ensures all teams operate within a common environment. Every component in Potion is self-contained, versioned, and isolated, thanks to strict adherence to Docker and UV-based Python environments. This architecture eliminates the common pitfalls of dependency conflicts and environment drift, which are especially prevalent in scientific workflows.
Modular components: Turning research into reusable building blocks
At DeepLife, we think of software components in a similar way to how molecular biologists think of functional domains: small, specialized units with clearly defined inputs and outputs. Rather than building end-to-end workflows as monoliths, we compose them from independently developed and tested components.
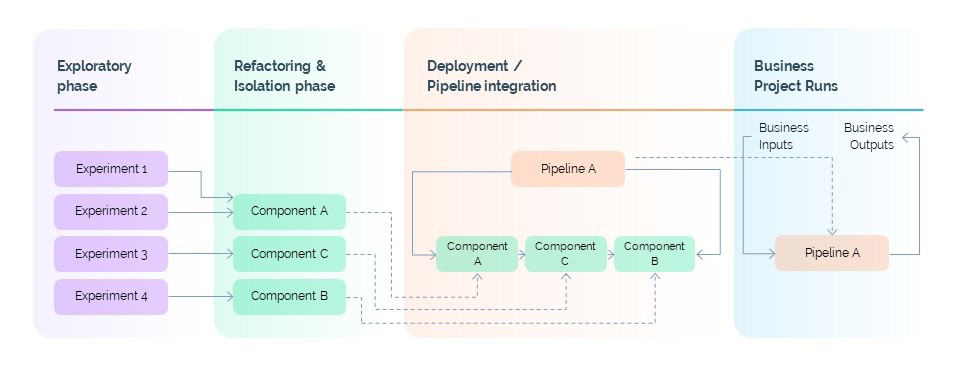
Each component adheres to the Single Responsibility Principle. One might handle quality control for a single-cell RNA-seq dataset. Another might perform dimensionality reduction. A third might execute clustering. These components can be independently developed, reviewed, and versioned - but they can also be easily assembled into more complex workflows through configuration.
This modular architecture delivers two major benefits. First, it accelerates experimentation: researchers can rapidly swap components in and out to evaluate new approaches. Second, it promotes reuse: a well-built component developed for one project can serve as the foundation for many others.
This isn’t a theoretical ideal - it’s how we work every day. In one recent project, a researcher prototyped a new analytical method in a notebook on Monday. By Wednesday, the logic was refactored into three production-ready components. By Friday, those components were deployed into a full pipeline and reused in two other projects. That velocity is only possible because of Potion’s structure.
Documentation that’s automatic, searchable, and transparent
As with most scientific work, documentation is both vital and fragile. Without clear documentation, projects become opaque, onboarding becomes painful, and reuse becomes impossible. But maintaining documentation manually is a burden - one that often falls by the wayside in high-velocity research environments.
To solve this, we’ve built a fully automated documentation system that operates alongside our codebase. Using the mkdocs-monorepo-plugin developed by Spotify, we consolidate all documentation across our monorepo into a single searchable site.
This system composes Markdown files, README documents, and Jupyter notebooks into a unified portal that mirrors the structure of the codebase. As researchers create new components or pipelines, their documentation is automatically indexed and exposed through our internal web application - without requiring any extra effort.
The result is a continuously updated knowledge graph that reflects the real state of the platform. It makes it possible to find, understand, and reuse components instantly. It also reduces onboarding time for new contributors, who can quickly navigate existing projects, see how components are connected, and identify what can be reused or extended.
In practice, this has created a culture of visibility and accessibility. New projects begin with discovery, not duplication. Researchers start from a place of context, not from scratch.
Integrated MLOps and CI/CD: From notebook to production in a smooth way
Bridging the gap between prototype and production requires more than clean code - it requires infrastructure. DeepLife’s internal platform automates this transition through a modern MLOps stack built directly into Potion.
Every component and pipeline is continuously tested using a combination of Pytest, Ruff, and Mypy. When a pull request is opened, our CI/CD pipelines - managed through GitHub Actions - validate the code, build container images, and deploy them to our Kubernetes cluster using Helm.
For researchers, this process is invisible and effortless. They focus on science; the platform handles the rest. This includes infrastructure provisioning, runtime validation, and environment configuration. When a component is ready, it can be deployed to production in minutes.
We also use MLFlow to track experiments and model performance, ensuring that decision-making is backed by transparent and auditable metrics. The integration with Potion allows us to trace every model back to its input data, training configuration, and component version.
Infrastructure that enables autonomy, not complexity
Beneath the surface, our infrastructure is built to be robust, scalable, and cost-effective. But more importantly, it is built to be invisible. We believe that researchers shouldn’t have to think about Kubernetes, YAML files, or autoscaling policies. They should think about data, models, and results.
To support this, we’ve invested in a Kubernetes-based platform that includes:
- Karpenter for dynamic autoscaling of compute resources
- Argo Workflows for orchestrating reproducible multi-step pipelines
- Nextflow for scalable bioinformatics workflows
- Devpod for launching cloud-based development environments that mirror production
All of this is provisioned through Terraform and managed centrally, so that researchers can deploy production-grade pipelines using a single command. Role-based access controls and namespace isolation ensure that security and governance never become bottlenecks.
This infrastructure empowers our team to move fast without breaking things or needing to understand the full depth of the systems powering their work.
Real-world impact: harnessing scalable and reproducible components for drug repositioning
We use Potion to support our own drug repurposing pipeline, leveraging existing components to expedite computation, quality control and reporting processes. By deploying standardized components as reproducible and scalable containers, we can quickly identify datasets, build atlases and identify condition-specific signatures that are used as input to our causal analysis methods.
Having those steps implemented as Potion components enables a plug-and-play approach to analysis, combining the power of modern pipeline frameworks with high-quality reproducible code. This transforms state-of-the-art methods into easy-to-use functions that can be easily swapped to address the specificities of the diverse biological data we are analyzing. Consequently, analysts at DeepLife are empowered to focus on interpretation rather than implementation, thereby maximizing the future positive impact our work will have for patients.
Building the future of biotech engineering
At DeepLife, we believe that research and production should not be separate concerns. They belong on a single continuum - supported by tools, practices, and infrastructure that respect the needs of both worlds.
Potion embodies this philosophy. It empowers researchers to remain agile and curious, while enabling engineers to build with precision and confidence. By investing in automation, modularity, and documentation, we’ve created an R&D platform that turns ideas into impact - quickly, reliably, and at scale.
This vision reflects more than a technical architecture; it’s rooted in the deeper principles of reproducible science and responsible engineering. As noted by the Stanford Encyclopedia of Philosophy, true reproducibility requires both the replication of results and the transparency of method—something Potion achieves through standardized interfaces, tracked experiments, and environment encapsulation along their reproducibility and portability..
Our approach also aligns with the design science research paradigm, where software components are treated as carefully designed, testable artifacts with measurable utility.
In many ways, Potion reflects the spirit of the Slow Science movement: fast to deploy, but built for thoughtful iteration, reusability, and long-term clarity. It resists the short-termism often seen in academic research or startup engineering, favoring a foundation that is rigorous, auditable, and scalable.
We’re not just imagining the future of biotech R&D. We’re engineering it - one well-scaffolded, reproducible component at a time.
Interested in how our approach can accelerate your projects?
Explore collaboration opportunities with DeepLife






