.png)
Scalable data architecture for biomedical discovery
Abstract
At DeepLife, our mission is to accelerate the discovery of new therapies through drug repurposing, leveraging large-scale biomedical data, bioinformatics pipelines, machine learning, and AI. Achieving this requires a data platform that is scalable, reproducible, and flexible enough to handle heterogeneous data from research publications, clinical studies, experimental results, and more.
In this blog post, we explain DeepLife’s modular infrastructure: from orchestrated pipelines using Nextflow, Kubeflow, and Argo Workflows, to a serverless analytics layer built on AWS S3, Glue, and Athena, and a centralized metadata and model tracking system powered by OpenMetadata and MLflow. This architecture allows our teams to seamlessly process, query, and analyze complex biomedical data at scale - supporting everything from alignment and annotation to AI model training and hypothesis testing.
We also explore how our evolving platform is addressing critical challenges around data changes (observability), making sure our results are reproducible, managing access securely (governance), and improving collaboration across different teams. As we continue to build a robust discovery engine for therapeutic innovation, DeepLife’s data infrastructure plays a key role in transforming raw data into actionable scientific insights faster and more reliably.
How we use data at DeepLife
At DeepLife, data is at the core of everything we do. From researchers analyzing the latest datasets on irritable bowel syndrome to large-scale bioinformatics workflows uncovering new therapeutic candidates for arthritis, data fuels our scientific discovery and innovation. It is not only a strategic asset but also a foundation upon which our research, bioinformatics and machine learning pipelines are built. To amplify the impact of our data across projects and teams, we apply FAIR principles to ensure it remains well managed, discoverable, interoperable and reusable:
- Findability: datasets and model artifacts are assigned globally unique persistent identifiers and richly described with metadata
- Accessibility: data and metadata are made available through standard protocols (e.g., AWS S3 APIs, Athena SQL), with secure authentication and authorization controls
- Interoperability: metadata employ community-recognized vocabularies and forms enabling integration across tools and domains
- Reusability: versioning, lineage and provenance information supports traceability and facilitates repurposing and validation
We collect data from diverse, often unstructured sources - scientific publications, biomedical repositories, public ontologies, and research datasets. This raw information is transformed through carefully designed pipelines into actionable outputs, such as ranked candidates for drug repurposing. Ensuring that this process is reproducible, scalable, and secure is essential to maintaining scientific rigor and operational efficiency.
The data we handle mirrors the complexity of biology itself. It includes:
- Massive biological files like single-cell RNA-seq datasets
- Heterogeneous formats with partially structured or unnormalized tabular data
- Experimental records that are incomplete or inconsistently annotated
- Multiple versions of datasets and models that evolve over time
To meet these challenges, we’ve built a robust, modular data platform that combines domain-specific pipeline orchestration tools, serverless querying and metadata discovery, and a powerful data catalog. Below, we’ll walk through the main components of our data architecture and how they work together.
Data architecture overview
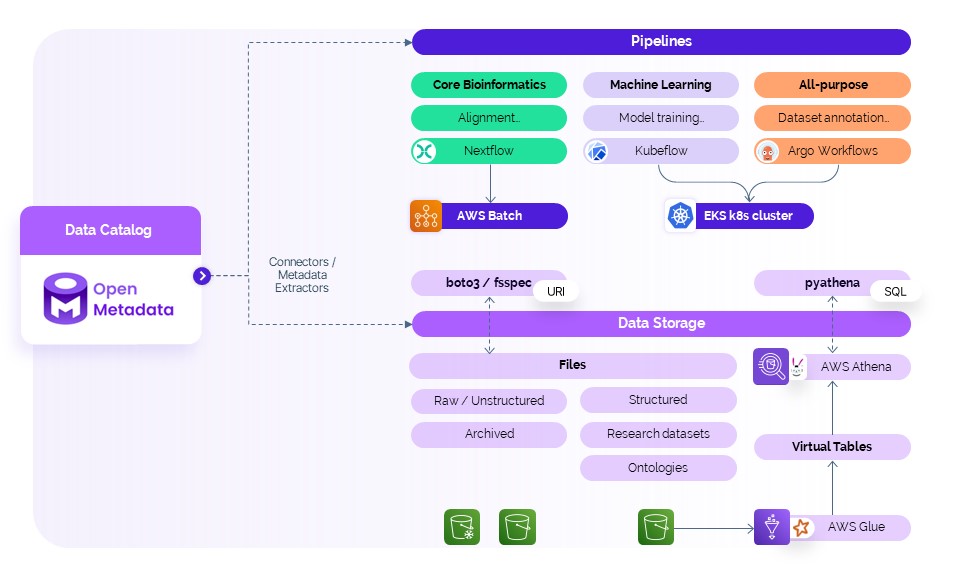
Modular pipeline architecture: tailoring workflows for bioinformatics, machine learning and data integration
DeepLife’s computational workflows are structured into three main types of pipelines, each designed for different tasks. This modular approach allows us to tailor infrastructure and execution strategies to the specific requirements of each task, ensuring scalability, efficiency, and reproducibility across our data lifecycle.
Core bioinformatics (genomic data processing)
Our core bioinformatics pipelines handle genomic data processing tasks such as read alignment, variant calling, or cell annotation. These are high-throughput, compute-intensive jobs with well-established steps, making them an ideal fit for Nextflow, a domain-specific language and workflow engine widely adopted in the bioinformatics community. We run these pipelines on AWS Batch, which allows us to dynamically scale compute resources based on demand, optimizing both performance and cost.
Machine learning (model training and evaluation)
The machine learning pipelines focus on model training, evaluation, and inference - key components underpinning our AI-driven insights. For these pipelines, we leverage Kubeflow, a platform designed specifically for machine learning workflows on Kubernetes. Running on our Amazon EKS (Elastic Kubernetes Service) cluster, Kubeflow provides a robust environment for orchestrating training jobs, managing experiment metadata, and integrating seamlessly with our ML ops tooling.
General purpose (data preparation and integration)
We maintain a category of all-purpose pipelines designed to support general tasks such as data annotation, preprocessing, and integration (ETL) workflows that fall outside the strict boundaries of bioinformatics or machine learning. For these, we use Argo Workflows, a lightweight and flexible Kubernetes-native workflow engine. Like Kubeflow, Argo runs on our EKS cluster and allows us to quickly spin up reproducible, containerized pipelines for a wide variety of custom data operations.
Our three-tiered pipeline architecture ensures that DeepLife’s platform remains adaptable to the diverse and evolving needs of biomedical data science, while keeping infrastructure aligned with each workload’s specific demands.
Effective and scalable data storage and access with AWS services
Managing and accessing data efficiently is central to the design of DeepLife’s platform. We work with many types of data - structured, semi-structured and unstructured - and primarily store it on Amazon S3. S3 gives us a reliable and cost-effective place to keep everything from genomic FASTQ files, H5AD files, to annotated CSVs and Parquet datasets. When accessing these files programmatically, we use the tool fsspec, enabling seamless integration with our pipelines and internal tools, regardless of whether the data resides locally or in cloud storage like S3. This allows us to simplify data access patterns across environments and build scalable workflows in a flexible way without needing to hardcode file system logic.
To provide a single point of access for our structured and semi-structured data, we’ve adopted AWS Athena, a serverless query engine that allows us to run SQL queries directly on S3-stored data. This means we don’t have to move data into a centralized data warehouse. With a Python library called PyAthena, we can execute complex queries from within our Python components, enabling us to join, filter, and aggregate data across various formats without extra effort. This setup eliminates the need for complex integration (ETL) processes, significantly reducing both development time and infrastructure maintenance. To complement Athena and automate schema discovery, we use AWS Glue crawlers.
Together, Athena and Glue provide fast, scalable analysis without the complexity of managing traditional databases, and they keep our data well organized and easy to find. Under the hood, Athena is powered by Trino (formerly PrestoSQL), a high-performance distributed SQL query engine designed for interactive analytics on large datasets. It enables us to run SQL queries directly on data stored in S3 without moving or loading it into a database, dramatically reducing setup time and cost. Meanwhile, Glue uses Apache Spark as its execution engine, allowing it to scale ETL and data transformation tasks horizontally, making it ideal for preprocessing complex, semi-structured, or large-scale datasets if needed.
For our machine learning workflows, we choose MLflow to manage and store all of our trained models. MLflow provides a centralized platform for tracking experiments, logging model parameters and metrics, and registering models for deployment. Models are stored in a structured, version-controlled format, often alongside artifacts and metadata, making it easy to reproduce training runs and monitor performance over time. This integration helps keep our machine learning pipelines transparent, reproducible, and production-ready.
Centralizing data discovery: DeepLife’s unified catalog
At DeepLife, we believe that data accessibility and transparency are key to driving efficient collaboration and generating insights across teams. That’s why we’ve built our data catalog around OpenMetadata, an open-source metadata platform designed to unify and govern modern data stacks. OpenMetadata brings together all types of data assets - such as files stored in S3, tables in Athena, machine learning models and pipeline metadata - into a single centralized place.
Whether new data or models are created, OpenMetadata automatically collects and organizes their metadata using built-in connectors with AWS Glue, Athena, and MLflow. This means that as soon as a new dataset, table, or model becomes available, it is cataloged with rich metadata and made instantly discoverable to everyone in the company. Team members can search and browse assets by name, tag, owner, or schema, significantly reducing the time spent hunting down the right data.
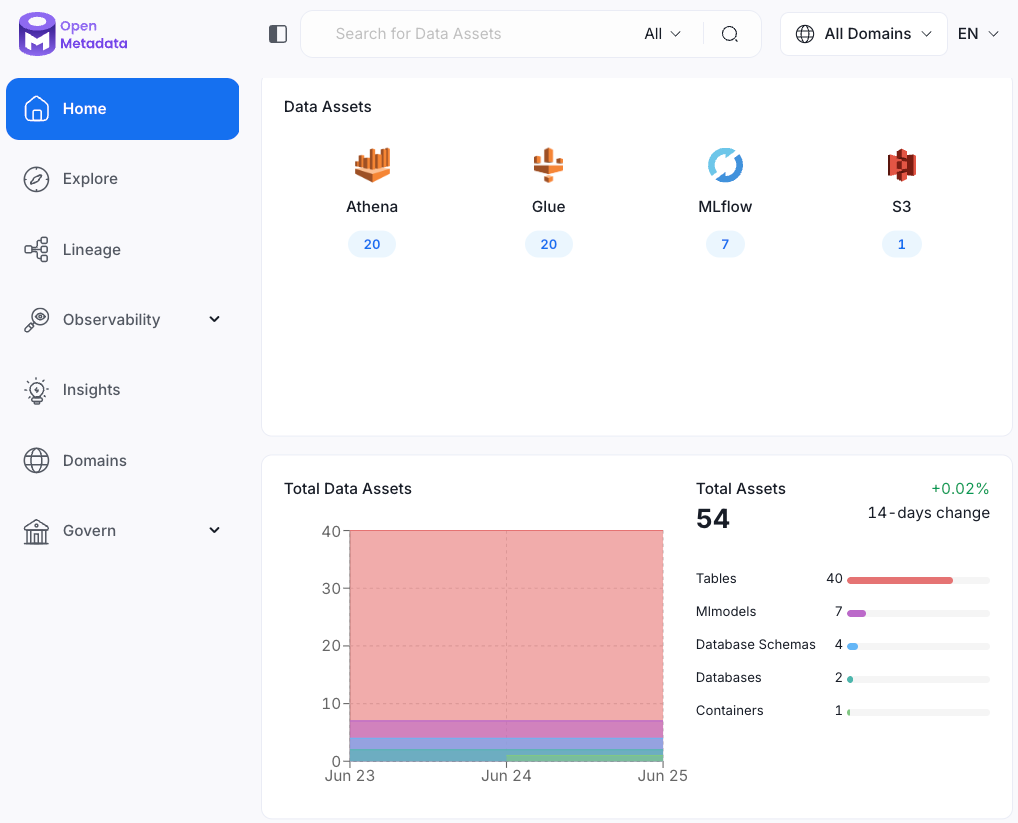
A key advantage of this data catalog approach is the ability to clearly see how data moves and changes across the organization. Users can view lineage graphs, see how datasets are used across pipelines, track model dependencies, and assess data quality metrics in one unified interface. This helps teams understand how data flows across the organization, identify potential issues early, and build with confidence.
OpenMetadata also provides an API, which allows us to integrate metadata programmatically into our workflows. Our teams can query the catalog to dynamically validate data availability before launching a pipeline, automate documentation, or trigger notifications when certain datasets change.
Next steps in advancing data quality, reproducibility and security for scalable drug discovery
As DeepLife’s platform continues to evolve, so does the scale, diversity, and strategic importance of the data we manage. Our current architecture - spanning automated pipelines, cloud analytics, model tracking, and a unified metadata catalog - provides a strong foundation. But to push the boundaries of translational science even further, we are actively expanding our capabilities in several key directions.
First, we’re boosting data observability and quality automation. With AWS Glue and OpenMetadata, we already track data structure and flow. Next, we’re integrating automated checks to spot potential anomalies early, ensuring our biomedical data stays accurate and reliable for making predictions.
As DeepLife grows, we’re enhancing our approach to versioning and reproducibility by linking models closely with their source data and pipeline settings with MLflow. This ensures full traceability of experimental results all the way back to the raw data. Alongside this, we’re strengthening data security by combining OpenMetadata’s role-based permissions in concert with AWS IAM policies to protect sensitive data while enabling collaboration in a safe environment.
Ultimately, our goal is to transform DeepLife’s data infrastructure into a powerful engine for generating and validating hypotheses, accelerating the discovery of repurposable drugs with greater accuracy and safety. By continuing to invest in automation, governance, and metadata-driven development, we are building more than just a data system - we are creating a discovery platform that will reshape the future of how therapeutics are found.






