Benchmarking interactomes: quality over quantity in molecular networks
Abstract
In the realm of molecular biology, the construction of interactomes - comprehensive maps of molecular interactions within a cell - has been pivotal for understanding complex biological systems. They are particularly useful for translating fundamental biological knowledge into biotechnological and pharmaceutical insights for target identification, biomarker discovery and drug repurposing. Traditionally, the emphasis has been on expanding these networks by increasing the number of interactions, operating under the assumption that a larger network equates to a more accurate representation of cellular processes. However, recent findings from DeepLife challenge this notion, suggesting that the quality and specificity of interactions are more critical than sheer quantity.
The DeepLife interactome: a paradigm shift
DeepLife's interactome offers a meticulously curated multi-omics interaction network that prioritizes interaction quality, directionality, and directness. Unlike other networks, it distinguishes between directed interactions, which clarify effector-affected relationships, and direct interactions, which involve physical contact. This careful curation ensures the integrity of the network, paving the way for more accurate analyses of cellular processes in drug target identification and other network analyses for biology.
The interactome can also be tailored to specific cell types, which is crucial because protein interactions vary substantially across different cell types and tissues. This specificity shapes network structure and coordination of cellular processes, including phenotypes and disease states.
Recent advances in computational models integrate protein expression and interaction data to predict these cell-type-specific interactions, enabling the discovery of novel targets unique to particular cell types. Focusing on cell-type specificity also helps unravel complex signaling pathways, where receptor-effector relationships vary and dictate diverse cellular responses. DeepLife’s cell-type-specific interactomes reflect the dynamic and context-dependent nature of protein networks, improving precision in biological insights and therapeutic development.
Method: interactome construction and validation
The distinction between directionality and directness is of high importance to this work. In a binary interaction, directionality is information about which of the two molecules is the source, performing a modification, and the target, being modified as a result of the interaction. Directness refers to whether the interaction is due to a direct contact between the two partners or is mediated by unknown intermediary molecules.
With interaction directionality and directness in mind, we surveyed available molecular interaction databases. A series of manually curated databases was identified, with an emphasis on causality and curation processes that purposely avoid indirect relationships. We then carefully selected relevant subsets of the identified databases and ran an in-house curation process, which is proprietary to DeepLife. Its purpose was to prune interactions that were not deemed reliable enough by our team of expert molecular biologists.
Benchmarking interactomes: quality over quantity
To assess the efficacy of DeepLife's interactome, we conducted a benchmarking study comparing it to other widely used interactomes, including those from the Barabási research group (which mostly contains indirect interactions) and the STRING database (which has many indirect interactions). We focused the evaluation on how accurately these networks could identify drug targets through perturbational experiments.
In these experiments, scientists disable (knock out) a gene in cells and measure which genes change their activity as a result, creating the resulting differentially expressed genes (DEG) profile. The idea is that in most diseases, a small initial change (perturbation) in a few genes leads to changes in many genes downstream. If a method can correctly identify the knock-out gene from the genes that changed, it’s likely good at finding the initially perturbed genes involved in diseases. These genes are seen as good potential drug target candidates as they address the root cause of diseases rather than their symptoms.
Practically, we mapped the knocked-out gene and the DEGs from the perturbational experiments to their corresponding nodes on the interactome. Then, we ranked all the genes in the network by how close they were to the changed genes. The lower the rank of the knockout gene node, the closer it was to the DEGs compared to other interactome nodes. We repeated this across 350 different perturbation experiments, the average rank of which can be found on the graph below. The lower the average knockout gene rank, the better a given interactome was deemed at recovering perturbations and performing target identification.
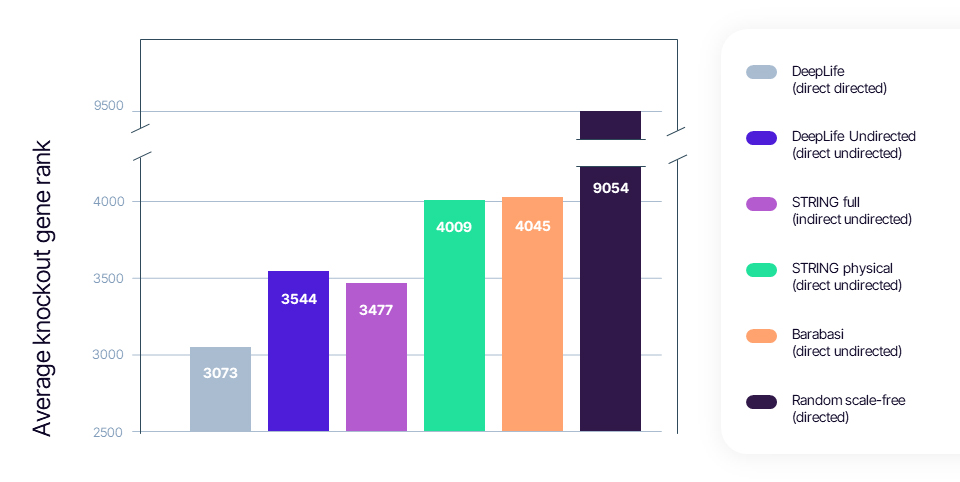
The results were telling:
- DeepLife's interactome: Demonstrated superior performance in drug target identification tasks, underscoring the value of focusing on interaction directionality and directness.
- Barabási's interactome: Yielded accuracy comparable to the STRING physical network, indicating that merely increasing the number of interactions does not necessarily enhance network performance.
- STRING network: The full STRING network, which contains a large portion of indirect interactions, performed worse than the DeepLife interactome, highlighting the importance of direct interactions in network efficacy.
These findings suggest that a more extensive network with numerous interactions does not inherently lead to better biological insights. Instead, the emphasis should be on the quality and specificity of the interactions included.
Implications for future research
The benchmarking results from DeepLife's interactome highlight a crucial consideration for researchers: the construction of interactomes should prioritize the accuracy and relevance of interactions over their sheer number. This approach not only enhances the network's utility in tasks like drug target identification but also provides a more faithful representation of cellular processes.
As the field advances, integrating high-quality, directionally and directly curated interactions will be essential for developing interactomes that truly reflect the intricate nature of biological systems. DeepLife's work serves as a compelling example of how focusing on interaction quality can lead to more meaningful and actionable biological insights.
References
- DeepLife Documentation. (n.d.). About The Interactome. Retrieved from https://deeplifeai.github.io/molecular_interaction_data/
- Stojmirović, A., & Yu, Y.-K. (2011). ppiTrim: Constructing non-redundant and up-to-date interactomes. arXiv preprint arXiv:1103.1402. Retrieved from https://arxiv.org/abs/1103.1402
- Huttlin, E. L., et al. (2021). Dual Proteome-scale Networks Reveal Cell-specific Remodeling of the Human Interactome. Cell, 184(11), 3022–3040.e28. Retrieved from https://pmc.ncbi.nlm.nih.gov/articles/PMC8165030/
Extracted from article originally published in the Book of Abstracts of the Complex Networks 2023 conference, pages 554-556.
Empower your strategy with quality interactomes
Connect with us to explore how insights from DeepLife's interactomes can enhance your approach.






